As tumor treatment enters the precision era, how to predict clinical drug efficacy for patients has become a key issue. Patient-derived organoids (PDOs) have huge potential in preserving tumor characteristics and drug efficacy. However, organoid culture and drug efficacy testing cycles are long and costs are high, and existing data volumes are insufficient to support deep learning models. How to fully develop and utilize the limited but high-quality organoid high-fidelity data, combined with large-scale cell line data foundations, to achieve more accurate and scalable clinical drug efficacy prediction models has become an important challenge.
Recently, the team of Prof. Zhao Bing from our Institute/School of Basic Medical Sciences/The First Affiliated Hospital of Nanchang University published a research paper titled “PharmaFormer predicts clinical drug responses through transfer learning guided by patient derived organoid” in the journal npj Precision Oncology. This study reported a model named PharmaFormer based on Transformer architecture and transfer learning, which achieved high-precision clinical drug efficacy prediction by combining PDOs and cell line data.

The core innovation of PharmaFormer lies in its unique three-stage transfer learning strategy. The model first utilizes gene expression data from over 900 cell lines and dose-response curves of over 100 drugs from the GDSC database for pre-training, capturing the complex patterns of gene-drug interactions. Subsequently, it is fine-tuned via limited but highly faithful organoid drug efficacy data (such as colon cancer and bladder cancer organoids), significantly enhancing clinical applicability. The model architecture ingeniously fuses gene expression and drug molecular structure: the gene feature extractor adopts dual linear layers to process RNA-seq data, while the drug feature extractor utilizes Byte Pair Encoding (BPE) to parse SMILES strings. These features are integrated by the Transformer encoder, outputting precise drug efficacy predictions.
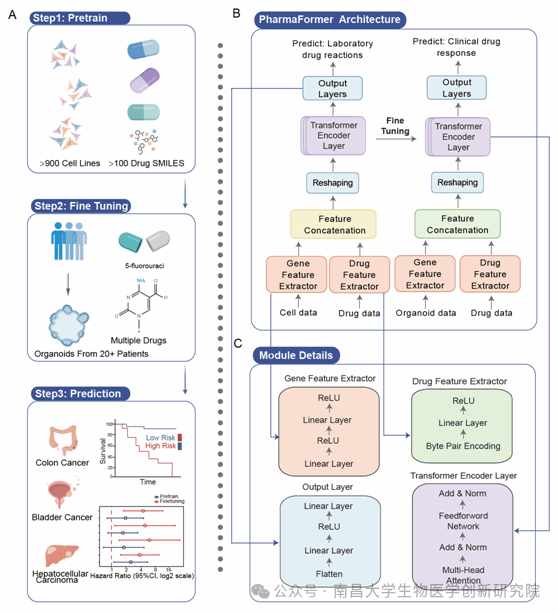
In performance validation, PharmaFormer demonstrated significant advantages. Compared with classic machine learning models, its Pearson correlation coefficient in the pre-training stage far exceeded traditional machine learning methods such as support vector machines and random forests. More crucially, the fine-tuned model achieved a qualitative leap in clinical prediction and was successfully validated in multiple independent cohorts. In patients with colon cancer, bladder cancer, and liver cancer, the fine-tuned version of PharmaFormer accurately distinguished drug responders from non-responders. Kaplan-Meier analysis showed that for colon cancer patients receiving 5-fluorouracil or oxaliplatin treatment, the survival rate of the high-risk group classified by the fine-tuned model was significantly reduced; similar results were reproduced in the bladder cancer cohort treated with gemcitabine/cisplatin. This breakthrough stems from the biological fidelity of organoid data—when the model was fine-tuned only with cell lines, the prediction effect was far inferior to the organoid fine-tuned version.

PharmaFormer is expected to fully excavate the important value of organoid high-fidelity data through the cross-integration of AI and organoids, while simultaneously compensating for the temporary shortcomings of long culture cycles and high costs, opening a new path for precision treatment.
It is worth mentioning that "Interdisciplinary Precision and Intelligent Medicine" has been included in Nanchang University's "15th Five-Year" key discipline construction, with "Organoids and New Drug Creation" as one of the important directions. The FDA also proposed an animal experiment replacement plan in 2025, explicitly identifying Organoids + AI as an important tool/method.
For this study, Zhou Yuru, a doctoral student at the School of Basic Medical Sciences of Nanchang University, is the first author, and Prof. Zhao Bing and Dr. Cheng Minzhang from our Institute/School of Basic Medical Sciences/The First Affiliated Hospital of Nanchang University are co-corresponding authors.
